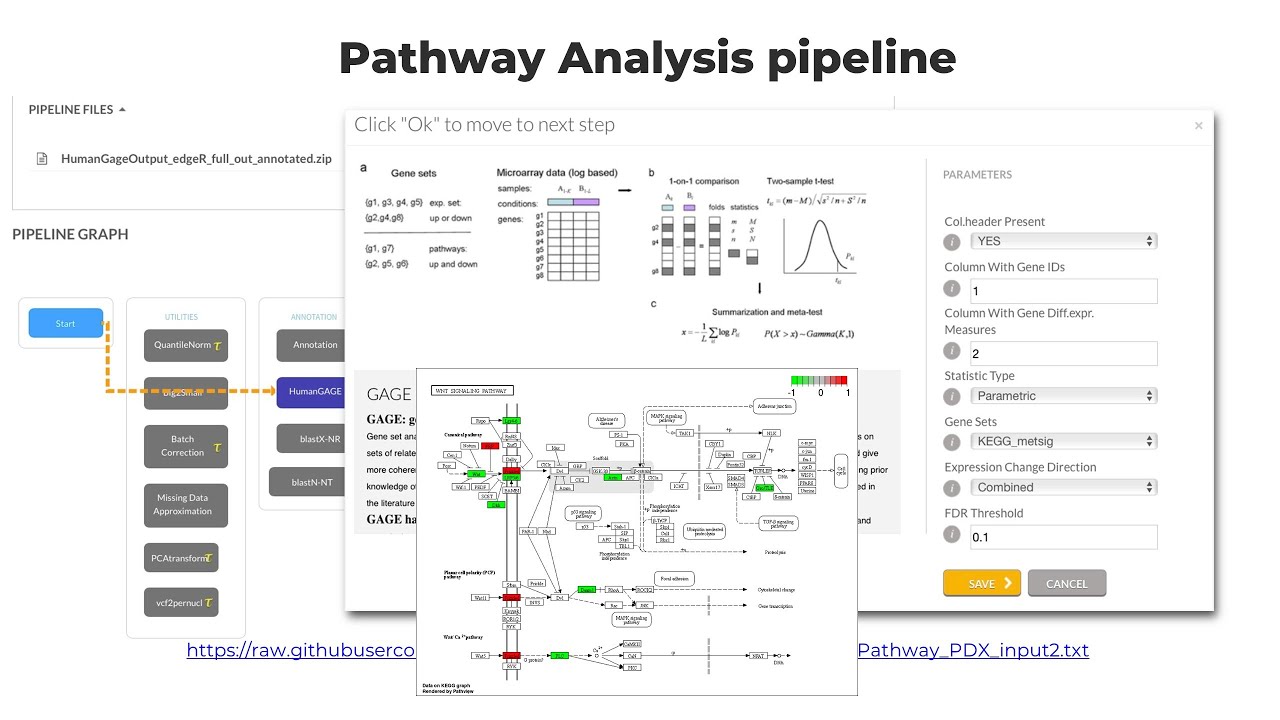
From the DESeq2 and GSEA analysis module, we can get important information about their biological implications. Besides, if you perform simply T-test or Use EdgeR, you will not get information regarding their annotation & biological implication in GO terms or plathways. So, what can we do in such scenarios ? In Response to this, we can perform gene annotation and pathway analysis independently using the Annotation and the HumanGAGE pathway analysis modules (also integrated on the T-Bioinfo server).
The annotation module allows us to annotate genes, i.e. Gene Symbol, Gene Description, Entrez Gene ID, ensembl transcript and gene ID based on the Ensemble IDs. Besides, we can also find important GO terms associated with each individual gene in the form of Table.
To learn the logic behind each step involved in the Annotation & Pathways analysis Pipeline, visit: https://learn.omicslogic.com/Learn/course-5-transcriptomics/lesson/09-t2-differential-gene-expression-and-gene-enrichment-analysis.
After completing the pipeline, you will get results in the zipped folder. You need to download the zipped folder. In the folder, you will find images for each pathway and a.txt file, which provide you with a pathway ID with corresponding P-value and P-adjusted value which indicates whether your list of genes are significantly enriched in the pathway or not.